Background: Monosomy 7/Del 7 (-7) or its long arm (del(7q)) is one of the most common cytogenetic abnormalities in pediatric and adult myeloid malignancies, particularly in adverse-risk acute myeloid leukemias (AMLs). In general, (-7) is associated with poor response to induction chemotherapy (PMID 12393746). At the same time, not all patients fare poorly so the ability to identify responders and non-responders remains a high priority.
Aim: To predict the response for induction chemotherapy in AML patients with (-7) and identify novel genomic signatures of response and resistance.
Methods: Genomic data from 13 consecutive patients with (-7) were analyzed using the Cellworks Omics Biology Model (CBM) to generate patient-specific protein network models. All data was anonymized, de-identified and exempt from IRB review. For each model, disease simulations were performed and patients were segregated into HOXA-upregulated and HOXA-downregulated cohorts based on the simulation levels of HOXA5 and HOXA9. Digital drug simulations for induction chemotherapy were accomplished by measuring the impact of drug effect on a cell growth score, a composite of cell proliferation, viability and apoptosis indices. Each patient-specific model was analyzed to identify mechanisms underlying treatment outcomes.
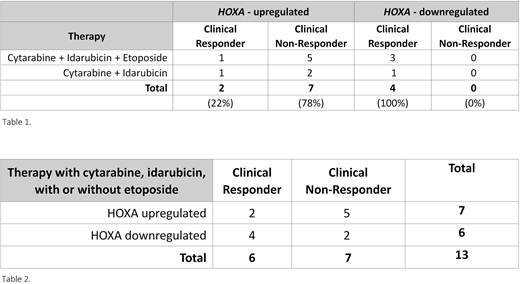
Results: 7/13 (54%) of (-7) patients failed to achieve remission after induction chemotherapy (Table 1) which highlighted that (-7) alone does not confer resistance to chemotherapy. CBM identified other genomic alterations that determine chemotherapy response, including DNA repair deficiency genes, mismatch repair (MMR), and homologous recombination repair (HRR) genes. DNA methylation and histone methylation (H3K27me) impacting HOXA gene expression, mainly HOXA5 and HOXA9, were identified as upstream regulators of DNA repair genes. Loss or reduced levels of EZH2 is associated with lower H3K27 methylation and thereby higher expression of HOXA5 and HOXA9 gene targets. High active levels of HOXA correlated with low rates of successful remission induction (22%, n=7) and lower activity levels of HOXA correlated with successful induction chemotherapy (100%, n=4) (Table 1).
CBM identified that (-7) results in a decreased expression of EZH2, CARD11, EIF3, PMS2, HUS1, KMT2C (MLL3), CDK5 and IKZF1 genes. Since EZH2 abnormalities alone are not implicated in poor prognosis for AML patients, we sought other aberrations that prevent HOXA upregulation. Using CBM, we identified multiple accompanying aberrations which regulate HOXA genes. Deletions of KAT6A, ASXL1, DNMT3B, DNMT3L genes and high KMT2A-partial tandem duplication correspond to HOXA-upregulation whereas EED amplification, gain of function mutations in DNMT3A, mutations in IDH1/2, MYC and KAT6A amplification and KDM4A deletion result in HOXA-downregulation.
Conclusion: Alterations of chromatin regulation have consequences for transcription factors that regulate expression of DNA repair genes. Under conditions where DNA repair is enhanced, induction chemotherapy was 78% less likely to effect remission in (-7) AML patients undergoing induction chemotherapy. Loss of H3K27 methylation associated with loss of PRC2 function by any means resulted in HOXA-upregulation and upregulation of DNA repair genes induced resistance to induction therapy. On the other hand, CBM analysis identified genetic signatures associated with a 100% remission rate from AML induction therapy despite the presence of (-7). Generation of H3K27me caused by PRC2 activation resulting from numerous mechanisms led to HOXA-downregulation and 100% response to induction therapy.
Stratification of patients harboring (-7) by HOXA biomarker analysis could inform treatment planning, avoid drug-related adverse events and reduce treatment costs after validation with a larger prospective dataset.
Castro:Cellworks Group Inc: Consultancy. Watson:Cellworks Group Inc.: Consultancy, Membership on an entity's Board of Directors or advisory committees; Cellmax Life Inc.: Consultancy, Membership on an entity's Board of Directors or advisory committees; Mercy Bioanalytics, Inc.: Consultancy, Membership on an entity's Board of Directors or advisory committees; SEER Biosciences, Inc.: Consultancy, Membership on an entity's Board of Directors or advisory committees; BioAI Health Inc.: Consultancy, Membership on an entity's Board of Directors or advisory committees. Kumar:Cellworks Research India Private Limited: Current Employment. Nair:Cellworks Research India Private Limited: Current Employment. Grover:Cellworks Research India Private Limited: Current Employment. Sahu:Cellworks Research India Private Limited: Current Employment. Mohapatra:Cellworks Research India Private Limited: Current Employment. G:Cellworks Research India Private Limited: Current Employment. Agarwal:Cellworks Research India Private Limited: Current Employment. Suseela:Cellworks Research India Private Limited: Current Employment. Ganesh:Cellworks Research India Private Limited: Current Employment. Sauban:Cellworks Research India Private Limited: Current Employment. Kumar:Cellworks Research India Private Limited: Current Employment. Raman:Cellworks Research India Private Limited: Current Employment. Singh:Cellworks Research India Private Limited: Current Employment. Basu:Cellworks Research India Private Limited: Current Employment. Lunkad:Cellworks Research India Private Limited: Current Employment. Mundkur:Cellworks Group Inc.: Current Employment. Macpherson:Cellworks Group Inc.: Current Employment. Kapoor:Cellworks Research India Private Limited: Current Employment. Howard:Servier: Consultancy, Other: Speaker; Boston Scientific: Consultancy; Sanofi: Consultancy, Other: Speaker; EUSA Pharma: Consultancy; Cellworks: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.